System¶
General overview¶
Data defining the system, like crystallographic data, is stored in groups within system. If more than one system are to be stored in the same system group, then each should go to its own subgroup. The choice of name for the subgroup is left to the user, with the restriction that it cannot be any of the names already in use in these specifications. If only one system is to be specified, then it can be stored directly in the system group or in its own subgroup.
The group must have the following attributes:
- system_name
- number_of_physical_dimensions
- dimension_types
- lattice_vectors
- embedded_system
- number_of_species
- number_of_sites
The group may have the following optional attributes:
- number_of_symmetry_operations
- symmorphic
- time_reversal_symmetry
- spacegroup_3D_number
- bulk_regions_for_semi_infinite_dimension
The group must contain the following datasets:
- species_at_sites
The group must contain at least one of the following datasets:
- cartesian_site_positions
- fractional_site_positions
The group must contain at least one of the following datasets:
- species_names
- chemical_symbols
- atomic_numbers
The group may contain the following optional datasets:
- reduced_symmetry_matrices
- reduced_symmetry_translations
- number_of_species_at_site
- concentration_of_species_at_site
- local_rotations
- magnetic_moments
- site_regions
- cell_in_host
- site_in_host
- forces
- stress_tensor
Detailed description of variables¶
General variables¶
These variables convey the most basic information regarding the geometry of the system. They are all mandatory.
- system_name: attribute, char(80) Specifies the name of the system. This information is stored for debugging or visualization purposes.
- number_of_physical_dimensions: attribute, unsigned int (always
3) The number of physical dimensions in space. Note that this is not the same as the number of periodic directions, which might be less than or equal to this number. - dimension_types: attribute, int
[number_of_physical_dimensions] (between
0and2) This is a list defining the periodicity of the system in each of the directions given by the lattice_vectors. Valid options are:0: The direction is non-periodic.1: The direction is periodic.2: The direction is semi-infinite. Only one direction can take this value; if it is present, then additional variables are required (see variables relating to a semi-infinite setup).
- embedded_system: attribute, char(3) (
yesorno) Is the system embedded into a host geometry? Ifyes, then additional variables are required, and the host geometry should be described in a separate group (see variables relating to an embedded system).
Variables relating to the cell¶
These variables define and describe properties of the unit cell. Only the first is mandatory. Note that the number of lattice vectors must be equal to the number of physical dimensions, even if some of these are non-periodic (see dimension_types). In this case, lattice vectors in non-periodic directions are not used, other than for defining fractional_site_positions; we suggest to set them either to an orthonormalized set or to a large box containing the molecule. The latter would be particularly useful for a periodic code reading in the geometry.
- lattice_vectors: attribute, double [number_of_physical_dimensions] [number_of_physical_dimensions] (dimensional variable: length) Holds the real-space lattice vectors (in Cartesian coordinates) of the simulation cell. The last (fastest) index runs over the x,y,z Cartesian coordinates, and the first index runs over the 3 lattice vectors.
- bulk_regions_for_semi_infinite_dimension: see variables relating to a semi-infinite setup
- stress_tensor: see variables relating to relaxation and MD
Variables relating to species¶
These variables define the available species (i.e., possible types of inequivalent sites). The species can be described in three different ways, at least one of which must be included; however, more than one might be necessary to provide a complete description.
- number_of_species: attribute, unsigned int The number of different species in the system.
- species_names: dataset, char(80) [number_of_species]
Descriptive name for each species. Could simply be equal to
chemical_symbols or contain extra information (e.g.,
Ga-semicore,C-1s-corehole,C-sp2,C1, etc.) - chemical_symbols: dataset, char(3) [number_of_species]
The chemical symbol for each species.
Xmay be used for a non-traditional atom (see atomic_numbers). - atomic_numbers: dataset, double [number_of_species] (dimensional variable: charge) The atomic number for each species. This could be non-integer for a number of reasons (e.g., a VCA atom), or zero (e.g., an empty site). In such cases we recommend using species_names to clarify the nature of the site.
Variables relating to sites¶
These variables define the position and attributes of each site in the unit cell. Only the first four are mandatory. Note that it is possible to define sites which are a statistical mixture of more than one species; the number of component species can be specified individually for each site. Some of the properties of the site relate to the site as a whole (i.e., its position), while others need to be specified for each component species (i.e., the magnetic moment).
- number_of_sites: attribute, unsigned int The number of sites in the unit cell.
- cartesian_site_positions: dataset, double [number_of_sites] [number_of_physical_dimensions] (dimensional variable: length) The position of each site in cartesian (absolute) coordinates.
- fractional_site_positions: dataset, double [number_of_sites] [number_of_physical_dimensions] The position of each site in fractional (reduced/crystallographic) coordinates.
- species_at_sites: dataset, unsigned int [number_of_sites]
[number_of_species_at_site(site_index)]
This variable defines the species at each site, according to the list
specified previously (see variables relating to species). If
[number_of_species_at_site(site_index)] is set to
1, the site is simply a single species; otherwise, it will be a mixture of more species. - number_of_species_at_site: dataset, unsigned int
[number_of_sites]
The number of component species for each site. If not present, it is
taken to be
1for all sites (i.e., no statistical mixing). - concentration_of_species_at_site: dataset, double [number_of_sites] [number_of_species_at_site(site_index)] The statistical concentration of each component species at each site. This variable needs to be present if number_of_species_at_site is present; otherwise, it is not used.
- local_rotations: dataset, double [number_of_sites] [number_of_physical_dimensions] [number_of_physical_dimensions] A rotation matrix defining the orientation of each site. If the rotation matrix only needs to be specified for some sites, the remaining sites should set it to the zero matrix (not the identity!)
- magnetic_moments: dataset, double [number_of_sites] [number_of_species_at_site(site_index)] [number_of_physical_dimensions] (dimensional variable: magnetic moment) The magnetic moment of each component at each site. If the magnitude is not important, we recommend to normalize the vector. Please remember that the Bohr magneton has a value of \(1/2\) in atomic units!
- site_regions: see variables relating to a semi-infinite setup
- cell_in_host: see variables relating to an embedded system
- site_in_host: see variables relating to an embedded system
- forces: see variables relating to relaxation and MD
Variables relating to spatial symmetry¶
The symmetry variables are optional. If the symmetry of the system is unknown, they should all be excluded. If the symmetry is to be specified, at least the first three need to be included.
- number_of_symmetry_operations: attribute, unsigned int The number of symmetry operations.
- reduced_symmetry_matrices: dataset, double [number_of_symmetry_operations] [number_of_physical_dimensions] [number_of_physical_dimensions] The transformation matrix in reduced coordinates and real space for each symmetry operation. For periodic crystals, these can be expressed purely in integers, but for arbitrary point groups, this is not possible.
- reduced_symmetry_translations: dataset, double [number_of_symmetry_operations] [number_of_physical_dimensions] The translation vector in reduced coordinates (without a factor of \(2 \pi\)) for each symmetry operation.
- spacegroup_3D_number: attribute, unsigned int (between
1and232) Specifies the International Union of Crystallography (IUC) number of the 3D space group that defines the symmetry group of the simulated physical system. - symmorphic: attribute, char(3) (
yesorno) Is the space group symmorphic? Set toyesif all translations are zero.
Variables relating to magnetic symmetry¶
These variables are optional. Further specifications may be needed for magnetic space groups and the action of symmetry operations on the magnetic moments.
- time_reversal_symmetry: attribute, char(3) (
yesorno) Is time-reversal symmetry present?
Variables relating to a semi-infinite setup¶
A semi-infinite setup is one in which a particular lattice direction (see dimension_types) is split into three regions: crystal 1, central region, crystal 2. Both crystals are semi-infinite and terminate at opposite ends of the central region. If this is the case, the additional variables listed below are needed. They define the unit cell of the two crystals, contained within the lattice vector of the whole system.
- bulk_regions_for_semi_infinite_dimension: attribute, double
[
2] (dimensional variable: length) The length of the lattice vector in the semi-infinite direction for the two crystals (see figure below). - site_regions: dataset, int [number_of_sites] (between
0and2) Each site in the system can either belong to the central region (0), or be part of the unit cell of crystal 1 (1) or crystal 2 (2).
Schematic of the semi-infinite setup.
The above figure shows a schematic of the semi-infinite setup. The lattice vectors of the cell are \(\left \{ \mathbf{a}_1, \mathbf{a}_2, \mathbf{a}_3 \right \}\) (defined in lattice_vectors), those of crystal 1 are \(\left \{ \mathbf{b}_1, \mathbf{b}_2, \mathbf{b}_3 \right \}\), and those of crystal 2 are \(\left \{ \mathbf{c}_1, \mathbf{c}_2, \mathbf{c}_3 \right \}\). It should be clear that \(\mathbf{c}_1 \equiv \mathbf{b}_1 \equiv \mathbf{a}_1\) and \(\mathbf{c}_2 \equiv \mathbf{b}_2 \equiv \mathbf{a}_2\), and so \(\left \{ \mathbf{b}_1, \mathbf{b}_2, \mathbf{c}_1, \mathbf{c}_2 \right \}\) need not be specified. The lattice vectors of the two crystals in the semi-infinite direction are defined as:
\(\mathbf{b}_3 = \alpha \mathbf{a}_3 / \left | \mathbf{a}_3 \right |\)
and
\(\mathbf{c}_3 = \beta \mathbf{a}_3 / \left | \mathbf{a}_3 \right |\);
bulk_regions_for_semi_infinite_dimension stores the values \(\alpha\) and \(\beta\).
Variables relating to an embedded system¶
If embedded_system is set to yes, the geometry described is
taken to be that of a finite region embedded into a larger host system.
In this case, two important things must be noted: Firstly, the embedded
geometry must be zero-dimensional (i.e., entirely non-periodic, with
dimension_types set to (0,0,0)). Secondly, a host
geometry must be specified in a separate group. This host geometry will
have embedded_system set to no, and has no restrictions in its
periodicity; it may even contain a semi-infinite dimension.
The additional variables listed below need to be specified in the embedded geometry. They relate each site of the embedded geometry to a site in a supercell of the host geometry.
- cell_in_host: dataset, int [number_of_sites]
[number_of_physical_dimensions]
The cell indices of the equivalent site in the host supercell. If the
site is one that does not exist in the host (i.e., for an
interstitial defect), the values are not referenced (we suggest
setting them to
0). If a direction is semi-infinite, the corresponding index will depend on which region the equivalent host site is in: if it is in the central region, the value must be0; if it is in one of the two crystal regions, the value must be greater than or equal to0, denoting the cell index of the semi-infinite crystal it belongs to. - site_in_host: dataset, unsigned int [number_of_sites]
(between
0and number_of_sites of the host geometry). The site index of the equivalent site in the host geometry (between1and number_of_sites specified in the host geometry). If the site is one that does not exist in the host, this should be indicated by setting the value to0.
Finally, it is important to note the behaviour of species_at_sites for an embedded geometry. The species defined for a site can either be identical to that of the equivalent host site, or different (e.g., for a substitutional defect). If a host site needs to be removed (e.g., for a vacancy), the site should be included in the embedded geometry, and the species should be set to an empty site (see atomic_numbers).
Variables relating to relaxation and MD¶
These variables are optional.
- forces: dataset, double [number_of_sites] [number_of_physical_dimensions] (dimensional variable: force) Forces on each site.
- stress_tensor: dataset, double [number_of_physical_dimensions] [number_of_physical_dimensions] (dimensional variable: pressure) Stress tensor. Express any relevant conventions here!
NOMAD Meta Info¶
The ESCDF specifications for the system group follow closely the section_system from the NOMAD Meta Info. There was a effort from both projects to keep the specifications fully compatible, so any changes in these specifications should be discussed and agreed with the NOMAD project.
The following list indicates the differences between the two specifications:
- NOMAD meta info uses booleans, while ESCDF uses a char(3) with yes and no as allowed values.
- NOMAD meta info uses SI units, while ESCDF allows for different unit systems with atomic units being the default.
- number_of_sites corresponds to number_of_atoms in NOMAD.
- cartesian_site_positions corresponds to atom_positions in NOMAD.
Examples¶
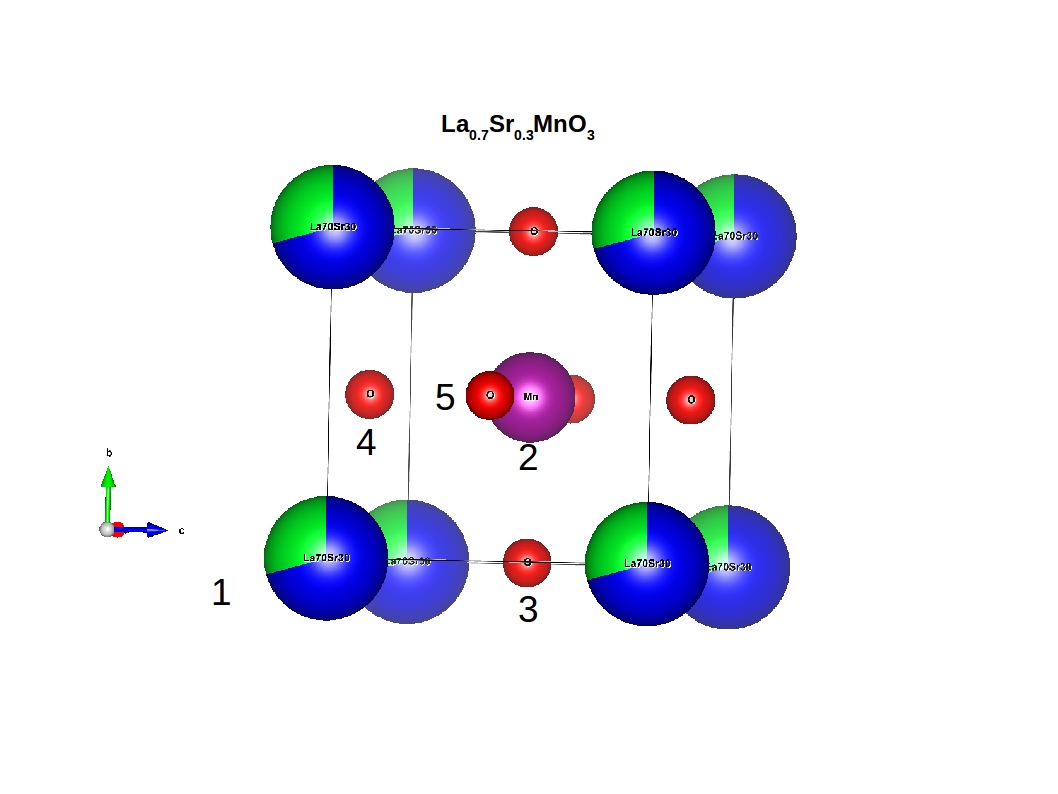
Example for partial occupations¶
In the case of partial occupations number of species on one site is not 1. Above we show example of LSMO in perovskite structure: number_of_sites=5 number_of_species=4 (La, Sr, O, Mn) having number_of_species_at_site[1]=2 with occupations concentration_of_species_at_site[1][1]=0.7 and concentration_of_species_at_site[1][2]=0.3